T-tests¶
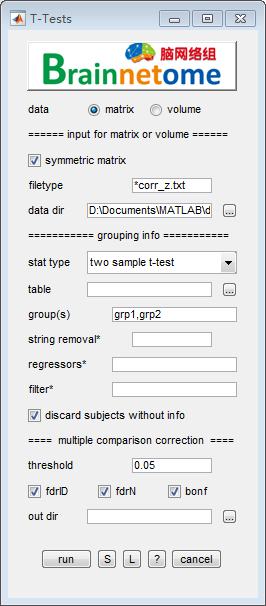
- data: matrix
- symmetric matrix: check to extract the upper right matrix’s elements, uncheck to extract all elements
- data: volume
- mask: could be whole brain mask or gray matter mask.
filetype: filetype
data dir: Input directories of matrices.
- grouping info:
stat type: one sample t-test, two sample t-test and paired t-test.
- groups:
- for two sample t-test, e.g. SZ,NC -> will do two-sample t-test for SZ and NC
- for paired t-test, e.g. SZ,NC -> will do paired t-test for SZ and NC
- for one sample t-test, e.g. SZ;NC -> will do one-sample t-test for both SZ and NC group
table: A comma-seperated values (csv) table, which is used for parsing subject names and covariates. The parsed names/ids will be matched to search results conducted with datadir and filetype. Before matching, BRANT will remove specified strings.
For one and two sample t-tests:
name group filter age subj1 SZ center1 28 subj2 SZ center1 27 subj3 NC center1 30 subj4 NC center2 25 For paired t-test, another column of paired_t_idx is required to specify paired subjects in each group:
name group filter age paired_t_idx subj1 stage1 center1 28 1 subj2 stage1 center1 27 2 subj3 stage2 center1 30 1 subj4 stage2 center2 25 2 string removal*: optional. remove strings from search results parsed by id index.
regressors*: optional. title of regressors which will be regressed out before statistical analysis. e.g. age
filter*: optional. use the control for subject in different state or center, fill in center1 here and subject4 won’t be included in the analysis.
discard subjects without info: when checked, if subjects’ information are not found in the table, a warning message will be shown; when unchecked, an error message will be shown.
- Multiple comparison correction methods (voxel-wise)
- threshold: the level of MULCC
- fdrID: false discovery rate (independent input)
- fdrN: false discovery rate (inputs not independent)
- bonf: Bonferroni correction for family wise error rate
out dir: output directory for saving results.
- Buttons:
- S: Save parameters of the current panel to a
*.matfile. The*.matcan be further loaded for the panel or be used in a script processing. - L: Load parameters from
*.matfor the current panel. - ?: Help information.
- S: Save parameters of the current panel to a